Professional Documents
Culture Documents
A Simple Genetic Algorithm: Appendix B
Uploaded by
cbqucbquOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
A Simple Genetic Algorithm: Appendix B
Uploaded by
cbqucbquCopyright:
Available Formats
Appendix B
A Simple Genetic Algorithm
The m-files given in the Appendices that follow can be downloaded from the
authors web site www.lar.ee.upatras.gr/reking
% Main_Program - ga.m
popsize=10; % Population Size
maxgen=50; % Number of Generations (iterations)
length=12; % Length of genotype (i.e. number of bits
in the binary array)
pcross=0.8; % Probability of Crossover
pm=0.01; % Probability of Mutation
bits=[length length];
vlb=[-1 -1];
vub=[1 1];
phen=init(vlb,vub,popsize,2); % Initialization of the
population of phenotypes
[gen, lchrom, coarse, nround] = encode(phen, vlb,
vub, bits); % Conversion of phenotypes to binary
string
[fitness, object]=score(phen,popsize); % Evaluation
of Fitness & Objective Function
x=-1:0.1:1; % Display of the contour graph of the
objective function
y=-1:0.1:1;
[x1,y1]=meshgrid(x,y);
z=x1.^2+y1.^2-0.3*cos(3*pi*x1)-0.4*cos(4*pi*y1)+0.7;
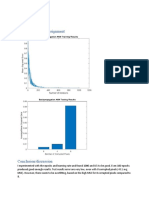
figure(1);
contour(x,y,z); 279
280 Appendix B
[best_obj(1), index] = min(object); % Store the best
candidate of the initial population
best_gen=gen(index,:);
best_phen=phen(index, :);
[worst_obj(1), index1] = max(object); % Store the
worst candidate of the initial population
worst_cur_gen=gen(index1);
worst_cur_phen=phen(index1);
avg_obj(1)=0; % Calculate the average performance of
the population
for k=1:popsize
avg_obj(1)=avg_obj(1)+object(k);
end;
avg_obj(1)=avg_obj(1)/popsize;
best_x(1)=best_phen(1);
best_y(1)=best_phen(2);
for i1=1:2
fprintf(1,'%f ',best_phen(1));
end;
fprintf('\n');
fprintf(1,'BEST : %f WORST : %f AVG : %f
\n',best_obj(1),worst_obj(1),avg_obj(1));
for i=1:maxgen % Start of the Genetic-Loop
newgen=reproduc(gen,fitness); % Reproduction
gen=mate(gen); % Mate two members of the
population
gen=xover(gen, pcross); % Crossover Operation
gen=mutate(gen, pm); % Mutation Operation
[phen, coa] = decode(gen, vlb, vub, bits); %
Decode the genotype of the new population
to phenotype
[fitness, object]=score(phen,popsize); %
Evaluation
of the Fitness & Objective Functions
[best_cur_obj, index] = min(object); % Store
the best candidate of the current population
best_cur_gen=gen(index, :);
best_cur_phen=phen(index, :);
[worst_obj(i+1), index1] = max(object); % Store
the worst candidate of the current population
worst_cur_gen=gen(index1);
worst_cur_phen=phen(index1);
avg_obj(i+1)=0; % Average performance of the
current population
for k=1:popsize
avg_obj(i+1)=avg_obj(i+1)+object(k);
Simple Genetic Algorithm 281
end;
avg_obj(i+1)=avg_obj(i+1)/popsize;
if(best_cur_obj > best_obj(i)) % Apply Elitist
Strategy
phen(index1,:) = best_phen;
gen(index1,:) = best_gen;
object(index1) = best_obj(i);
best_obj(i+1) = best_obj(i);
elseif(best_cur_obj <= best_obj(i))
best_phen = best_cur_phen;
best_gen = best_cur_gen;
best_obj(i+1) = best_cur_obj;
end;
best_x(i+1)=best_phen(1); % Display evolution
of the best solution on the contour graph
best_y(i+1)=best_phen(2);
hold;
line(best_x,best_y);
for i1=1:2
fprintf(1,'%f ',best_phen(i1));
end;
fprintf(1,'---> %f\n',best_obj(i+1));
fprintf('\n');
fprintf(1,'BEST : %f WORST : %f AVG : %f
\n',best_obj(i+1),worst_obj(i+1),avg_obj(i+1));
end
xx=1:maxgen+1; % Display evolution of objective
functions for the worst, average and best
solutions
figure(2);
plot(xx,best_obj,xx,worst_obj,xx,avg_obj);
grid;
% File init.m - This function creates a random
population
function phen=init(vlb,vub, siz, sea)
for i=1:siz
phen(i,:)=(vub-vlb).*rand(1, sea) + vlb;
end
% File score.m - This function computes the fitness
and the objective function values of a population
function [fitness, object]=score(phen, popsize)
for i=1:popsize
282 Appendix B
object(i)=phen(i,1)^2+phen(i,2)^2-
0.3*cos(3*pi*phen(i,1))-
0.4*cos(4*pi*phen(i,2))+0.7;
fitness(i)=1/(object(i)+1);
end
The following m-files called by the main
program can be downloaded directly from the
MATHWORKS web site www.mathworks.com and
bear the indication:
% Copyright (c) 1993 by the MathWorks, Inc.
% Andrew Potvin 1-10-93.
% File encode.m - This function converts a variable
from
real to binary
function [gen,lchrom,coarse,nround] =
encode(x,vlb,vub,bits)
lchrom = sum(bits);
coarse = (vub-vlb)./((2.^bits)-1);
[x_row,x_col] = size(x);
gen = [];
if ~isempty(x),
temp = (x-ones(x_row,1)*vlb)./ ...
(ones(x_row,1)*coarse);
b10 = round(temp);
nround = find(b10-temp>1e-4);
gen = b10to2(b10,bits);
end
% File reproduc.m - This function selects individuals
in accordance to their fitness
function [new_gen,selected] = repro-
duc(old_gen,fitness)
norm_fit = fitness/sum(fitness);
selected = rand(size(fitness));
sum_fit = 0;
for i=1:length(fitness),
sum_fit = sum_fit + norm_fit(i);
index = find(selected<sum_fit);
selected(index) = i*ones(size(index));
end
new_gen = old_gen(selected,:);
% File mate.m - This function mates two members of
the population
Simple Genetic Algorithm 283
function [new_gen,mating] = mate(old_gen)
[junk,mating] = sort(rand(size(old_gen,1),1));
new_gen = old_gen(mating,:);
% File xover.m - This function performs the Crossover
operation
function [new_gen,sites] = xover(old_gen,Pc)
lchrom = size(old_gen,2);
sites = ceil(rand(size(old_gen,1)/2,1)*(lchrom-1));
sites = sites.*(rand(size(sites))<Pc);
for i = 1:length(sites);
new_gen([2*i-1 2*i],:) =
old_gen([2*i-12*i],1:sites(i)) ...
old_gen([2*i 2*i-1],sites(i)+1:lchrom)];
end
% File mutate.m - This function performs the Mutation
operation
function [new_gen,mutated] = mutate(old_gen,Pm)
mutated = find(rand(size(old_gen))<Pm);
new_gen = old_gen;
new_gen(mutated) = 1-old_gen(mutated);
% File decode.m - This function coverts a variable
from binary to real
function [x,coarse] = decode(gen,vlb,vub,bits)
bit_count = 0;
two_pow = 2.^(0:max(bits))';
for i=1:length(bits),
pow_mat((1:bits(i))+bit_count,i) =
two_pow(bits(i):-1:1);
bit_count = bit_count + bits(i);
end
gen_row = size(gen,1);
coarse = (vub-vlb)./((2.^bits)-1);
inc = ones(gen_row,1)*coarse;
x = ones(gen_row,1)*vlb + (gen*pow_mat).*inc;
% File b10tob2 - This function converts a variable
from base 10 to base 2
function b2 = b10to2(b10,bits)
bit_count = 0;
b2_index = [];
284 Appendix B
bits_index = 1:length(bits);
for i=bits_index,
bit_count = bit_count + bits(i);
b2_index = [b2_index bit_count];
end
for i=1:max(bits),
r = rem(b10,2);
b2(:,b2_index) = r;
b10 = fix(b10/2);
tbe = find( all(b10==0) | (bits(bits_index)==i) );
if ~isempty(tbe),
b10(:,tbe) = [];
b2_index(tbe) = [];
bits_index(tbe) = [];
end
if isempty(bits_index),
return
end
b2_index = b2_index-1;
end
You might also like
- Advanced Optimization Using MATLAB by EndalewDocument22 pagesAdvanced Optimization Using MATLAB by EndalewMerera TaresaNo ratings yet
- MATLAB Codes for Particle Swarm Optimization (PSO) AlgorithmDocument4 pagesMATLAB Codes for Particle Swarm Optimization (PSO) AlgorithmIzza AnshoryNo ratings yet
- Codes in MATLAB For Particle Swarm Optimization: March 2016Document4 pagesCodes in MATLAB For Particle Swarm Optimization: March 20162k19ee113 HitenNo ratings yet
- Codes in MATLAB For Particle Swarm Optimization: March 2016Document4 pagesCodes in MATLAB For Particle Swarm Optimization: March 2016Ashish MaheshwariNo ratings yet
- CodeDocument5 pagesCodeAbhay SharmaNo ratings yet
- Primenjeni Algoritmi Prvi KolokvijumDocument6 pagesPrimenjeni Algoritmi Prvi KolokvijumLazar ĐerićNo ratings yet
- PSO CodesDocument4 pagesPSO CodesSama TaleeNo ratings yet
- Codes in MATLAB For Particle Swarm Optimization: March 2016Document4 pagesCodes in MATLAB For Particle Swarm Optimization: March 2016sindhuja selvamNo ratings yet
- MATLAB Codes for Particle Swarm Optimization PSODocument4 pagesMATLAB Codes for Particle Swarm Optimization PSOGanesh C JNo ratings yet
- Appendix B MATLAB® PROGRAMS PDFDocument16 pagesAppendix B MATLAB® PROGRAMS PDFcassindromeNo ratings yet
- Tutorial Questions On Fuzzy Logic 1. A Triangular Member Function Shown in Figure Is To Be Made in MATLAB. Write A Function. Answer: CodeDocument13 pagesTutorial Questions On Fuzzy Logic 1. A Triangular Member Function Shown in Figure Is To Be Made in MATLAB. Write A Function. Answer: Codekiran preethiNo ratings yet
- Function To Generate PN SequenceDocument2 pagesFunction To Generate PN SequenceFetsum LakewNo ratings yet
- Contoh 1 Dynamic ProgrammingDocument6 pagesContoh 1 Dynamic ProgrammingDic alqolbNo ratings yet
- GA Code in MATLABDocument2 pagesGA Code in MATLABismail adeizaNo ratings yet
- Rosenbrock Function Optimization Using Gradient DescentDocument27 pagesRosenbrock Function Optimization Using Gradient DescentlarasmoyoNo ratings yet
- MATLAB Codes for Particle Swarm Optimization (PSO) Algorithm <40Document4 pagesMATLAB Codes for Particle Swarm Optimization (PSO) Algorithm <40palpandian.mNo ratings yet
- PSO Codes MatlabDocument4 pagesPSO Codes MatlabJosemarPereiradaSilvaNo ratings yet
- Misdayanti Praktikum 5Document13 pagesMisdayanti Praktikum 5Nurul HusnaNo ratings yet
- Matlab-Lecture 9 2018Document13 pagesMatlab-Lecture 9 2018berk1aslan2No ratings yet
- Multidimensional Arrays in MatlabDocument16 pagesMultidimensional Arrays in MatlabmanishtopsecretsNo ratings yet
- Implementation in Matlab of Differential EvolutionDocument26 pagesImplementation in Matlab of Differential EvolutionJuan Alex Arequipa ChecaNo ratings yet
- BaiTapLonGeneticAlgorithm_2019Document6 pagesBaiTapLonGeneticAlgorithm_2019Nhật TàiNo ratings yet
- Function SoalDocument8 pagesFunction SoalSiti FauziahNo ratings yet
- MOPSODocument7 pagesMOPSOkennyh1No ratings yet
- Codes in MATLAB For Particle Swarm Optimization: FunctionDocument3 pagesCodes in MATLAB For Particle Swarm Optimization: FunctionKorbi AmiraNo ratings yet
- Example From SlidesDocument17 pagesExample From SlidesShintaNo ratings yet
- CBNST Lab FileDocument20 pagesCBNST Lab FileMohit Singh0% (1)
- Plane Stress, Constant Strain Triangle, Matlab ScriptDocument10 pagesPlane Stress, Constant Strain Triangle, Matlab ScriptrohitNo ratings yet
- Back Propagation - Machine LearningDocument8 pagesBack Propagation - Machine LearningEvelyn MahasinNo ratings yet
- Pso Code MatlabDocument7 pagesPso Code Matlab08mpe026100% (2)
- ML Lab ManualDocument37 pagesML Lab Manualapekshapandekar01100% (1)
- Find A Real Root of The Equation F (X) X: ProblemDocument7 pagesFind A Real Root of The Equation F (X) X: ProblemMd. Faisal ChowdhuryNo ratings yet
- Bai Tap 2Document21 pagesBai Tap 2Bơ BòNo ratings yet
- PL IiDocument43 pagesPL IiRohan 7No ratings yet
- AppendixDocument13 pagesAppendixDania AlishaNo ratings yet
- DSP HW 1Document8 pagesDSP HW 1aboodm20thNo ratings yet
- 1 Homework 2: 1.1 Large Scale Data Analysis / Aalto University, Spring 2023Document12 pages1 Homework 2: 1.1 Large Scale Data Analysis / Aalto University, Spring 2023Ido AkovNo ratings yet
- Assignment ReportDocument35 pagesAssignment ReportTuong Nguyen Minh NhatNo ratings yet
- Introduction To Matlab: Pantech SolutionsDocument130 pagesIntroduction To Matlab: Pantech SolutionsMuzamil AhamedNo ratings yet
- Hyperparameter Tuning in XGBoost Using Genetic AlgorithmDocument11 pagesHyperparameter Tuning in XGBoost Using Genetic AlgorithmDubstar Prince Kaushik100% (1)
- Cuckoo Search With Levy FlightDocument4 pagesCuckoo Search With Levy Flightashikhmd4467No ratings yet
- Job Shop Scheduling Using Genetic AlgorithmDocument9 pagesJob Shop Scheduling Using Genetic Algorithmanurag 8055No ratings yet
- Comparing test errors of AdaBoost and Bagging classifiersDocument12 pagesComparing test errors of AdaBoost and Bagging classifierssurbhiNo ratings yet
- Machine Learning Lab Manual: Implement Backpropagation AlgorithmDocument13 pagesMachine Learning Lab Manual: Implement Backpropagation AlgorithmKumaraswamy AnnamNo ratings yet
- Pattern Recognition: Exercise 1Document5 pagesPattern Recognition: Exercise 1Ţoca CosminNo ratings yet
- An Introduction To An Introduction To Optimization Optimization Using Using Evolutionary Algorithms Evolutionary AlgorithmsDocument45 pagesAn Introduction To An Introduction To Optimization Optimization Using Using Evolutionary Algorithms Evolutionary AlgorithmsSai Naga Sri HarshaNo ratings yet
- Attachment 1Document2 pagesAttachment 1McPhillips GieggerNo ratings yet
- Numerical Methods and Linear Algebra Programs in CDocument20 pagesNumerical Methods and Linear Algebra Programs in CKESHAV SUDIALNo ratings yet
- Scoa CodesDocument9 pagesScoa CodeskpNo ratings yet
- TP 03: Technique D'optimisation PSODocument3 pagesTP 03: Technique D'optimisation PSOCEM Yelle N centreNo ratings yet
- Numerical Methods ProgrammingDocument9 pagesNumerical Methods ProgrammingMudit SrivastavaNo ratings yet
- PSODocument4 pagesPSOdinhsacNo ratings yet
- Exe 1Document13 pagesExe 1jaya pandiNo ratings yet
- GA MainDocument5 pagesGA Main05-CSEB2-3No ratings yet
- DSA LAB Quick Sort and Binary Search Lab ManualDocument51 pagesDSA LAB Quick Sort and Binary Search Lab ManualRichieRosewallNo ratings yet
- FOR Function 1 Question 1 Bisection Method: f1 @ (X) 2-X+log (X)Document9 pagesFOR Function 1 Question 1 Bisection Method: f1 @ (X) 2-X+log (X)superman1234No ratings yet
- Function: 'Buffalo (I,:) BP - K (I) 'Document5 pagesFunction: 'Buffalo (I,:) BP - K (I) 'Atul NarkhedeNo ratings yet
- Ebook - Driving Better Decision Making With Big Data - LNS ResearchDocument36 pagesEbook - Driving Better Decision Making With Big Data - LNS ResearchcbqucbquNo ratings yet
- User-Guide-Pioneer-Child Seat PDFDocument100 pagesUser-Guide-Pioneer-Child Seat PDFcbqucbquNo ratings yet
- Be A Field Operator in Hysys-Based Ots and Oculus Rift Virtual RealityDocument13 pagesBe A Field Operator in Hysys-Based Ots and Oculus Rift Virtual RealitycbqucbquNo ratings yet
- Aiche 2016 - Dynamic Simulation For Apc Total-InprocessDocument29 pagesAiche 2016 - Dynamic Simulation For Apc Total-InprocesscbqucbquNo ratings yet
- Ydstie PDFDocument26 pagesYdstie PDFcbqucbquNo ratings yet
- Ydstie PDFDocument26 pagesYdstie PDFcbqucbquNo ratings yet
- Ydstie PDFDocument26 pagesYdstie PDFcbqucbquNo ratings yet
- Ebook - Driving Better Decision Making With Big Data - LNS ResearchDocument36 pagesEbook - Driving Better Decision Making With Big Data - LNS ResearchcbqucbquNo ratings yet
- Ebook Edge Computing Stratus Special Edition PDFDocument35 pagesEbook Edge Computing Stratus Special Edition PDFTitipong PulbunrojNo ratings yet
- Grinding Mill Circuits - A Survey of Control and Economic ConcernsDocument6 pagesGrinding Mill Circuits - A Survey of Control and Economic ConcernscbqucbquNo ratings yet
- Ydstie PDFDocument26 pagesYdstie PDFcbqucbquNo ratings yet
- Ydstie PDFDocument26 pagesYdstie PDFcbqucbquNo ratings yet
- First-Principles Modeling For Optimization of Process OperationsDocument37 pagesFirst-Principles Modeling For Optimization of Process OperationscbqucbquNo ratings yet
- Two Decades After Vanishing, Her Daught... Speaking Spanish - The Washington PostDocument6 pagesTwo Decades After Vanishing, Her Daught... Speaking Spanish - The Washington PostcbqucbquNo ratings yet
- Edge Computing CE1812 120418 Stratus Final1Document39 pagesEdge Computing CE1812 120418 Stratus Final1cbqucbquNo ratings yet
- Ninja Master Prep Splash Guard Lid For QB900B QB900 16 Oz Cup Bowl - Kitchen & DiningDocument6 pagesNinja Master Prep Splash Guard Lid For QB900B QB900 16 Oz Cup Bowl - Kitchen & DiningcbqucbquNo ratings yet
- Ydstie PDFDocument26 pagesYdstie PDFcbqucbquNo ratings yet
- Ninja Master Prep Splash Guard Lid For QB900B QB900 16 Oz Cup Bowl - Kitchen & DiningDocument6 pagesNinja Master Prep Splash Guard Lid For QB900B QB900 16 Oz Cup Bowl - Kitchen & DiningcbqucbquNo ratings yet
- Introduction to Model Predictive Control (MPCDocument50 pagesIntroduction to Model Predictive Control (MPCcbqucbquNo ratings yet
- Progress ERTC03 R41Document11 pagesProgress ERTC03 R41cbqucbquNo ratings yet
- Economic MPC and Real-time Decision Making for Large-Scale HVAC Energy SystemsDocument59 pagesEconomic MPC and Real-time Decision Making for Large-Scale HVAC Energy SystemscbqucbquNo ratings yet
- Ydstie PDFDocument26 pagesYdstie PDFcbqucbquNo ratings yet
- Oil and Gas Offshore Pipeline Leak Detection System: A Feasibility StudyDocument8 pagesOil and Gas Offshore Pipeline Leak Detection System: A Feasibility StudycbqucbquNo ratings yet
- IDC Marketscape PTCDocument9 pagesIDC Marketscape PTCcbqucbquNo ratings yet
- Advanced Process Control Delivers Significant Economic Savings for Alumina PlantsDocument6 pagesAdvanced Process Control Delivers Significant Economic Savings for Alumina PlantscbqucbquNo ratings yet
- Plantwide Dynamic Simulators Luyben 2002Document448 pagesPlantwide Dynamic Simulators Luyben 2002cbqucbqu100% (1)
- Alumina APC Application at Alunorte (BRA) by Honeywell by 2Document5 pagesAlumina APC Application at Alunorte (BRA) by Honeywell by 2cbqucbquNo ratings yet
- Lu 1Document38 pagesLu 1cbqucbquNo ratings yet
- The Impact of Digit Alizat Ion On T He Fut Ure of Cont Rol and Operat IonsDocument39 pagesThe Impact of Digit Alizat Ion On T He Fut Ure of Cont Rol and Operat IonscbqucbquNo ratings yet
- Lu 1Document38 pagesLu 1cbqucbquNo ratings yet
- NewsDocument26 pagesNewsMaria Jose Soliz OportoNo ratings yet
- Lecture 2 Principle of EMRDocument33 pagesLecture 2 Principle of EMRizhar engkuNo ratings yet
- Advu en PDFDocument65 pagesAdvu en PDFGustavo Rodrigues de SouzaNo ratings yet
- MleplustutorialDocument13 pagesMleplustutorialvorge daoNo ratings yet
- ISCOM HT803 DatasheetDocument2 pagesISCOM HT803 Datasheetnmc79No ratings yet
- Excel Dynamic Arrays: Department Item Quantity Price Total $Document5 pagesExcel Dynamic Arrays: Department Item Quantity Price Total $Bilal Hussein SousNo ratings yet
- Curtis CatalogDocument9 pagesCurtis CatalogtharngalNo ratings yet
- Rso PDFDocument120 pagesRso PDFjohn shepardNo ratings yet
- HI-8592, HI-8593, HI-8594: Single-Rail ARINC 429 Differential Line DriverDocument14 pagesHI-8592, HI-8593, HI-8594: Single-Rail ARINC 429 Differential Line DriversameeppaiNo ratings yet
- 997-3 CIP Safety Adapter: Single Point Lesson (SPL) - Configure CIP Safety Adapter and A-B PLCDocument18 pages997-3 CIP Safety Adapter: Single Point Lesson (SPL) - Configure CIP Safety Adapter and A-B PLCTensaigaNo ratings yet
- DC Machines Chapter SummaryDocument14 pagesDC Machines Chapter SummaryMajad RazakNo ratings yet
- Network Layer: Computer Networking: A Top Down ApproachDocument83 pagesNetwork Layer: Computer Networking: A Top Down ApproachMuhammad Bin ShehzadNo ratings yet
- Design Plan: A Performance Task in GeometryDocument12 pagesDesign Plan: A Performance Task in GeometryRobert Ryan SantiagoNo ratings yet
- Potenciometro 15KDocument8 pagesPotenciometro 15Kra101208No ratings yet
- Product - 20V4000G24F 3B FODocument32 pagesProduct - 20V4000G24F 3B FOmohammed khadrNo ratings yet
- Erc111 DKRCC - Es.rl0.e3.02 520H8596Document24 pagesErc111 DKRCC - Es.rl0.e3.02 520H8596Miguel BascunanNo ratings yet
- Introduction To Curve FittingDocument10 pagesIntroduction To Curve FittingscjofyWFawlroa2r06YFVabfbajNo ratings yet
- Geophysical Report Megnatic SurveyDocument29 pagesGeophysical Report Megnatic SurveyShahzad KhanNo ratings yet
- Cantors Paradox PDFDocument16 pagesCantors Paradox PDFColectivo Utopía MoreliaNo ratings yet
- Handout 06 - Geothermometry PDFDocument7 pagesHandout 06 - Geothermometry PDFOg LocabaNo ratings yet
- QAF10A200S TheTimkenCompany 2DSalesDrawing 03 06 2023Document1 pageQAF10A200S TheTimkenCompany 2DSalesDrawing 03 06 2023LeroyNo ratings yet
- Sensors 22 09378 v2Document13 pagesSensors 22 09378 v2FahdNo ratings yet
- ENGG1330 2N Computer Programming I (20-21 Semester 2) Assignment 1Document5 pagesENGG1330 2N Computer Programming I (20-21 Semester 2) Assignment 1Fizza JafferyNo ratings yet
- Canalis KDP-KBA-KBB-KNA-KSA-20-1000A-2014Document324 pagesCanalis KDP-KBA-KBB-KNA-KSA-20-1000A-2014Rubén González CabreraNo ratings yet
- Simultaneous EquationsDocument11 pagesSimultaneous EquationsSaleena AurangzaibNo ratings yet
- MAINTAIN COOLANT LEVELDocument6 pagesMAINTAIN COOLANT LEVELAgustin BerriosNo ratings yet
- Technical Data: Pump NameDocument6 pagesTechnical Data: Pump Nameسمير البسيونىNo ratings yet
- 0001981572-JAR Resources in JNLP File Are Not Signed by Same CertificateDocument13 pages0001981572-JAR Resources in JNLP File Are Not Signed by Same CertificateAnonymous AZGp1KNo ratings yet
- Example 1 LS Dyna - Bullet Model SimulationDocument6 pagesExample 1 LS Dyna - Bullet Model Simulationsunil_vrvNo ratings yet
- Javascript Api: Requirements Concepts Tutorial Api ReferenceDocument88 pagesJavascript Api: Requirements Concepts Tutorial Api ReferenceAshish BansalNo ratings yet