Professional Documents
Culture Documents
Bioinformatics Lab 1
Uploaded by
Fiqa SuccessOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Bioinformatics Lab 1
Uploaded by
Fiqa SuccessCopyright:
Available Formats
CBE647 Bioinformatics Laboratory 1
The goal of this lab is to give you practical experience using the NCBI interface: how to navigate the website, how to perform basic and advanced searches. You will need to draw on what you learned in lectures, as well as the on-line help available to you through the website itself. Becoming experienced in using these sites will help you in the future as you try to identify novel genes or perform your own research. A. Finding Public Biological Databases The 2010 Database Issue of Nucleic Acids Research is the sixteenth in a series dedicated to factual biological databases. Such databases are an essential resource for working biologists and this compilation provides descriptions of the most important of these databases and serves to introduce newly compiled databases that provide specialist information in the biological area. NAR Online contains hotlinks to all of the databases in the compilation as well as brief summaries of their content. Go to the NAR (http://www.oxfordjournals.org/nar/database/a/) Visit databases of your own selection to see how these databases are accessed and what information is available. Include Genbank (Nucleotide Sequence Databases), KEGG (Metabolic Pathways), UniProtKB (Proteins), OMIM and GO (Gene Ontology) in your visit.
B. NCBI Entrez and Searching Biological Databases Problem: Triose Phosphate Isomerase We are going to investigate the human triose phosphate (or triosephosphate) isomerase 1 gene. This gene is responsible for the reaction that converts dihydroxyacetone phosphate to glyceraldehyde-3-phosphate in glycolysis. Glycolysis is the pathway in cells where a simple sugar (glucose) is transformed into two pyruvate molecules, which are then used to generate energy for the cell. Much is known about this gene, and when it is deficient, severe problems can occur. A loss of function mutation in this gene would be lethal. First, visit the NCBI website (http://ncbi.nlm.nih.gov/) and visit the All Databases page. 1. What would be a good search query to use for this gene that would specify both the name of the gene as well as the organism? For your answer here, dont worry about the difference between triose phosphate vs. triosephosphate; however, you may have to use both searches for the rest of the lab. Use this search query in the Gene, Protein, and Nucleotide sections of Entrez. You should observe different results in each, although they will contain much similar information.
2. What is the RefSeq accession number for this gene in the mRNA form and for the protein form? 3. On what chromosome is this gene found? 4. How many amino acids are in the protein chain? What are the first five? One of the useful abilities of Entrez is to cross reference recent publications that relate to this gene. A recent paper published implicates the triose phosphate isomerase protein in the disease Lupus. 5. Who were the three authors of this paper? What is this papers unique PubMed ID?
C. Determination of the Open Reading Frame (ORF) of the Hemoglobin Alpha 2 (HBA2) Gene. In this exercise you will learn how to determine an open reading frame (ORF) and determine the gene product of the ORF. A reading frame is a way of dividing the sequence of nucleotides in a nucleic acid (DNA or RNA) molecule into a set of consecutive, nonoverlapping triplets. Where these triplets equate to amino acids or stop signals during translation, they are called codons. A single strand of a nucleic acid molecule has a phosphoryl end, (called the 5-end) and a hydroxyl, or (3-end). These then define the 5'3' direction. An open reading frame (ORF) is the part of a reading frame that contains no stop codons. 1. Retrieve the alpha 2 globin mRNA sequence (NM_000517) from the GenBank database. Can you manually identify the Open Reading Frame (ORF), i.e., the coding sequence (e.g., in notepad or wordpad)? Proceed by determining the start and stop codons (use genetic code table). Note that the sequence contains triplets of nucleotides that are similar to the start/stop codons but which are not the true start and stop codons. Why is that? 2. Once you have determined the ORF of the HBA2 gene, translate the first 10 codons to the amino acid sequence (use genetic code table)
3. Are the ORF and the amino acid sequence confirmed by the NM_000517 annotation in the GenBank database?
4. For the automatic determination of putative ORFs you can also use the ORF finder at the NCBI site. Go to the ORF finder and copy/paste the NM_000517 sequence or just type in the accession code (the program is linked to the GenBank database). The results are the ORFs for all six reading frames. The longest ORF is most probably the frame that will be translated to the protein. By clicking on the largest ORF, the corresponding translation is given. Is this correct?
D. Sequence Extraction This part of the lab will guide you through the process of getting DNA sequences using the NCBI GeneBank database as a source. STEP 1 Go to the NCBI website STEP 2 Choose your search type (Nucleotide) and enter your search item in the box. Some examples of search items are: - ara h2 - opsins You will get a lot of results but for the purpose of this lab, find the following links and click on them: - Ara h2 ==> AY158467 - Opsins ==> NM_020061 STEP 3 The new page contains the DNA coding sequence for the proteins at the bottom, below Origin. Click and drag the cursor to highlight the entire sequence, right click the highlighted sequence and select copy to store it. - Ara h2 ==> from 1atggc to tactaa - Opsins ==> from cggctgccgt to ccaa *** Also copy this sequence and store in a .txt file. Remember to delete the numbers at the beginning of each row.*** STEP 4 Open the Expasy page to view the translation tool. This tool will do in seconds what will take you hours to do. It reads the codons in the sequence and translates them into proteins. STEP 5 Right click the cursor in the box below Please enter DNA and select paste to enter your gene sequence. To the right of Output format, select Includes nucleotide sequence from the drop-down menu and click Translate Sequence. Your results in the 5 3 Frame 1 should show the amino acid/ protein sequence of the gene in capital letters below the corresponding codons of the gene. Notice that: - Ara h2 ==> the gene starts with atg and the corresponding protein is M for methionine
- Opsins ==> the gene starts with cgg and the corresponding protein is R for arginine. The other frames translate the sequence but in an alternate direction from the 5 3 Frame 1 frame. STEP 6 Click on the 5 3 Frame 1 link to open another window with just the protein sequence. Click and drag the cursor to highlight the entire sequence, right click the highlighted sequence and select copy to store it. Now we are going to BLAST the sequence! BLAST is a tool that will match your sequence to any other similar sequences and give you a description of what your gene is/ does. Click http://www.ncbi.nlm.nih.gov/BLAST to open BLAST. STEP 7 Click protein blast and right click to paste your protein sequence into the large text box. Click BLAST. You have just asked the BLAST program to search the entire NCBI protein database for matches to your sequence. The BLAST results page can be a lot to take in, but the colour-coded graph shows the most similar sequence in red and other sequences that are less similar in magenta, green, blue and black. Under the graph, click on one of the links with a high score. On the resulting page, look for a DEFINITION or TITLE that will give you information about your gene sequence. For the examples we have been using, one of them is a peanut allergen and the other is an eye gene related to long-wave sensitivity and colour blindness. Can you tell which is which?
Submission Instructions: Please submit your individual laboratory report in the proper format by Wednesday (2nd April 2014) for EH222 8A or Thursday (3rd April 2014) for EH222 8B.
You might also like
- Film Interpretation and Reference RadiographsDocument7 pagesFilm Interpretation and Reference RadiographsEnrique Tavira67% (3)
- Imaging Anatomy Brain and Spine Osborn 1 Ed 2020 PDFDocument3,130 pagesImaging Anatomy Brain and Spine Osborn 1 Ed 2020 PDFthe gaangster100% (1)
- TDS-11SH Top Drive D392004689-MKT-001 Rev. 01Document2 pagesTDS-11SH Top Drive D392004689-MKT-001 Rev. 01Israel Medina100% (2)
- Fish Hematology in AquacultureDocument28 pagesFish Hematology in AquacultureEdwin A. Aguilar M.100% (1)
- Hydrotest Test FormatDocument27 pagesHydrotest Test FormatRähûl Prätäp SïnghNo ratings yet
- Multiple Sequence AlignmentDocument89 pagesMultiple Sequence AlignmentMeowNo ratings yet
- Molecular Identification, Systematics and Population Structure of ProkaryotesDocument328 pagesMolecular Identification, Systematics and Population Structure of Prokaryotesuriel franco100% (1)
- GYCDocument2 pagesGYCFiqa Success100% (1)
- Protein Synthesis (Gene Expression)Document63 pagesProtein Synthesis (Gene Expression)jaybeeclaireNo ratings yet
- Unit 2 Part 2: Transgenic Animals Transgenic Fish Animal As BioreactorsDocument43 pagesUnit 2 Part 2: Transgenic Animals Transgenic Fish Animal As BioreactorsPraveen KumarNo ratings yet
- Comparing DNA Sequences To Understand Evolutionary Relationships With BlastDocument3 pagesComparing DNA Sequences To Understand Evolutionary Relationships With Blastflyawayxx13No ratings yet
- Molecular Systematic of AnimalsDocument37 pagesMolecular Systematic of AnimalsMarlene Tubieros - InducilNo ratings yet
- Sequence AlignmentDocument92 pagesSequence AlignmentarsalanNo ratings yet
- Recombinant DNA Cloning TechnologyDocument71 pagesRecombinant DNA Cloning TechnologyRajkaran Moorthy100% (1)
- Advances in Zinc Finger Nuclease and Its ApplicationsDocument13 pagesAdvances in Zinc Finger Nuclease and Its ApplicationsFreddy Rodrigo Navarro GajardoNo ratings yet
- Bioinformatics Tutorial 2016Document28 pagesBioinformatics Tutorial 2016Rhie GieNo ratings yet
- Biodiversity (Semester 2 Matriculation)Document32 pagesBiodiversity (Semester 2 Matriculation)Sasikumar Kovalan100% (3)
- Chapter 7 Linkage, Recombination, and Eukaryotic Gene MappingDocument20 pagesChapter 7 Linkage, Recombination, and Eukaryotic Gene MappingSiamHashan100% (1)
- Types of Electrophoresis and Dna Fingerprinting B: 5,, ,: Y Group Lood Martos Panganiban TrangiaDocument73 pagesTypes of Electrophoresis and Dna Fingerprinting B: 5,, ,: Y Group Lood Martos Panganiban TrangiaJelsea AmarradorNo ratings yet
- Crispr Cas HajarDocument21 pagesCrispr Cas HajarHajira Fatima100% (1)
- Bioinformatics Exercises PrintDocument6 pagesBioinformatics Exercises Printalem010No ratings yet
- Bioinformatics AnswersDocument13 pagesBioinformatics AnswersPratibha Patil100% (1)
- Bioinformatics Database and ApplicationsDocument82 pagesBioinformatics Database and ApplicationsRekha Singh100% (2)
- Bioinformatics Lab 2Document9 pagesBioinformatics Lab 2NorAdilaMohdShahNo ratings yet
- PCR, Types and ApplicationDocument17 pagesPCR, Types and ApplicationRaj Kumar Soni100% (1)
- Molecular PhylogeneticsDocument4 pagesMolecular PhylogeneticsAb WassayNo ratings yet
- NCBI ResourcesDocument13 pagesNCBI ResourceshamzaloNo ratings yet
- Types of TransposonsDocument13 pagesTypes of Transposonsmozhi74826207No ratings yet
- An Introduction To Phylogenetic Analysis - Clemson UniversityDocument51 pagesAn Introduction To Phylogenetic Analysis - Clemson UniversityprasadburangeNo ratings yet
- Prepared By: Verna Jean M. Magdayao 3/Bsbiology/ADocument56 pagesPrepared By: Verna Jean M. Magdayao 3/Bsbiology/AKathleya PeñaNo ratings yet
- Unwrapping The StandardsDocument2 pagesUnwrapping The Standardsapi-254299227100% (1)
- Dadm Assesment #2: Akshat BansalDocument24 pagesDadm Assesment #2: Akshat BansalAkshatNo ratings yet
- Isolation of Genomic DNADocument8 pagesIsolation of Genomic DNAMadhuri HarshaNo ratings yet
- Spectrophotometry of DnaDocument7 pagesSpectrophotometry of DnaMel June FishNo ratings yet
- Lab 2 QuizDocument1 pageLab 2 QuizAAlbertoCNo ratings yet
- TVL-SMAW 12 - Week 4 - Lesson 1 - Concept of Welding Codes and StandardsDocument9 pagesTVL-SMAW 12 - Week 4 - Lesson 1 - Concept of Welding Codes and StandardsNelPalalonNo ratings yet
- STPM Bio P2 2011Document12 pagesSTPM Bio P2 2011Acyl Chloride HaripremNo ratings yet
- 02 Protein IsolationDocument14 pages02 Protein IsolationAntonio Calleja IINo ratings yet
- Industrial RevolutionDocument2 pagesIndustrial RevolutionDiana MariaNo ratings yet
- Unit 1: Structural GenomicsDocument4 pagesUnit 1: Structural GenomicsLavanya ReddyNo ratings yet
- Lect# Plant Molecular MarkersDocument23 pagesLect# Plant Molecular MarkersSohail Ahmed100% (1)
- BABS2213 TutorialDocument12 pagesBABS2213 TutorialAnusia ThevendaranNo ratings yet
- Lab 3 Standard Protein Assay LabDocument5 pagesLab 3 Standard Protein Assay LabMark WayneNo ratings yet
- Example of Lab Report PDFDocument15 pagesExample of Lab Report PDFAlejandra Acuña BalbuenaNo ratings yet
- Argus Molecular Docking 1Document5 pagesArgus Molecular Docking 1escherichioNo ratings yet
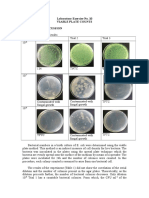
- Laboratory Exercise No. 10 Viable Plate Counts Results and DiscussionDocument3 pagesLaboratory Exercise No. 10 Viable Plate Counts Results and Discussionvanessa olga100% (1)
- Enzyme ImmobilizationDocument67 pagesEnzyme ImmobilizationBijayaKumarUpretyNo ratings yet
- Evaluating Cell Fractionation Procedures and Cytochemical TestsDocument6 pagesEvaluating Cell Fractionation Procedures and Cytochemical TestsJosh Zollman100% (1)
- Immobilization of EnzymesDocument21 pagesImmobilization of EnzymesSai SridharNo ratings yet
- Mitosis Lab 2Document4 pagesMitosis Lab 2api-235160519No ratings yet
- Genetic Engineering Applications in Animal BreedingDocument6 pagesGenetic Engineering Applications in Animal BreedingFadilla HadiwijayaNo ratings yet
- Benefits of BiopharmingDocument3 pagesBenefits of BiopharmingHarrcanaa Rajah50% (2)
- Crop ImprovementDocument14 pagesCrop ImprovementhappydaysvsNo ratings yet
- Classification of Plasmid Vectors Using Replication Origin, Selection Marker and Promoter As Criteria. WWW - Picb.ac - CNDocument5 pagesClassification of Plasmid Vectors Using Replication Origin, Selection Marker and Promoter As Criteria. WWW - Picb.ac - CNAhmad SolikinNo ratings yet
- Agrobacterium-Mediated Gene Transfer in Potato ForDocument19 pagesAgrobacterium-Mediated Gene Transfer in Potato FordelyadelzNo ratings yet
- Shotgun SequencingDocument29 pagesShotgun SequencingKhyati JoshiNo ratings yet
- Guide To Protein Purification, Volume 463Document6 pagesGuide To Protein Purification, Volume 463Dawlat SalamaNo ratings yet
- Serial Analysis of Gene Expression (SAGE)Document34 pagesSerial Analysis of Gene Expression (SAGE)Rohit PhalakNo ratings yet
- Isolation of DNA From Animal TissuesDocument10 pagesIsolation of DNA From Animal TissuesAnura BandaraNo ratings yet
- Practical 2 - 3 Monohybrid - Dihybrid CrossDocument5 pagesPractical 2 - 3 Monohybrid - Dihybrid CrossKhufaziera KharuddinNo ratings yet
- Animal Cell Culture - Part 1Document38 pagesAnimal Cell Culture - Part 1Subhi MishraNo ratings yet
- Seleksi Untuk Menyiapkan Bibit Jantan Domba Batur Di Kecamatan Batur, Kabupaten BanjarnegaraDocument24 pagesSeleksi Untuk Menyiapkan Bibit Jantan Domba Batur Di Kecamatan Batur, Kabupaten BanjarnegaraArifgiiNo ratings yet
- PFAM DatabaseDocument22 pagesPFAM DatabaseNadish KumarNo ratings yet
- Spinach Leaf Transformed Into Beating Human Heart TissueDocument13 pagesSpinach Leaf Transformed Into Beating Human Heart TissueDrishti MalhotraNo ratings yet
- SOLAF 2014 Biologi SPM Modul 3: JPN Perak 1Document7 pagesSOLAF 2014 Biologi SPM Modul 3: JPN Perak 1AZIANA YUSUFNo ratings yet
- Docking With ArgusLabDocument24 pagesDocking With ArgusLabDesmond MacLeod Carey100% (1)
- Mini PrepDocument6 pagesMini PrepWilson GomargaNo ratings yet
- Accessing Bibliographic DatabasesDocument25 pagesAccessing Bibliographic DatabasesNischith RkNo ratings yet
- A Method For Direct Harvest of Bacterial Cellulose Filaments During Continuous Cultivation of Acetobacter Xylinum PDFDocument5 pagesA Method For Direct Harvest of Bacterial Cellulose Filaments During Continuous Cultivation of Acetobacter Xylinum PDFFiqa SuccessNo ratings yet
- Optimization of Bacterial Cellulose Production FromDocument7 pagesOptimization of Bacterial Cellulose Production FromFiqa SuccessNo ratings yet
- PJFNS - 59 - 1 - 03 - Stasiak and BlazejakDocument8 pagesPJFNS - 59 - 1 - 03 - Stasiak and BlazejakFiqa SuccessNo ratings yet
- Design of Fill and Finish Facility For Active Pharmaceutical Ingredients (Api)Document20 pagesDesign of Fill and Finish Facility For Active Pharmaceutical Ingredients (Api)Fiqa SuccessNo ratings yet
- Biotechnology and Bioprocess Engineering: Instructions To AuthorsDocument4 pagesBiotechnology and Bioprocess Engineering: Instructions To AuthorsFiqa SuccessNo ratings yet
- ch1Document37 pagesch1Fiqa SuccessNo ratings yet
- Limitations in CelluloseDocument6 pagesLimitations in CelluloseShofiaNo ratings yet
- PJFNS - 59 - 1 - 03 - Stasiak and BlazejakDocument8 pagesPJFNS - 59 - 1 - 03 - Stasiak and BlazejakFiqa SuccessNo ratings yet
- Optimization of Bacterial Cellulose Production FromDocument7 pagesOptimization of Bacterial Cellulose Production FromFiqa SuccessNo ratings yet
- Optimization Production of Biocellulose by A. Xylinum Using RSMDocument7 pagesOptimization Production of Biocellulose by A. Xylinum Using RSMFiqa SuccessNo ratings yet
- Kipp 1995Document6 pagesKipp 1995Fiqa SuccessNo ratings yet
- Design of Fill and Finish Facility For Active Pharmaceutical Ingredients (Api)Document20 pagesDesign of Fill and Finish Facility For Active Pharmaceutical Ingredients (Api)Fiqa SuccessNo ratings yet
- Catalogue W03 Laboratory BioreactorsDocument8 pagesCatalogue W03 Laboratory BioreactorsFiqa SuccessNo ratings yet
- (54)Document8 pages(54)Fiqa SuccessNo ratings yet
- Bacteria CellulosaDocument10 pagesBacteria CellulosamelodicaNo ratings yet
- Cheng2009 PDFDocument10 pagesCheng2009 PDFFiqa SuccessNo ratings yet
- Effect of Structural Features On Enzyme Digestibility of Corn StoverDocument9 pagesEffect of Structural Features On Enzyme Digestibility of Corn StoverFiqa SuccessNo ratings yet
- PDF of CEE ReviewDocument5 pagesPDF of CEE ReviewFiqa SuccessNo ratings yet
- 08 NadiaDocument7 pages08 Nadiaelvina iskandarNo ratings yet
- (54)Document8 pages(54)Fiqa SuccessNo ratings yet
- Cannon 1991Document13 pagesCannon 1991Fiqa SuccessNo ratings yet
- From The Department of Biological Chemistry, Washington University School Medicine, St. LouisDocument12 pagesFrom The Department of Biological Chemistry, Washington University School Medicine, St. LouisFiqa SuccessNo ratings yet
- Sheet 1 Solution PDFDocument28 pagesSheet 1 Solution PDFVivek JoshiNo ratings yet
- Uhlin KiDocument131 pagesUhlin KiFiqa SuccessNo ratings yet
- Syafiqah ITC2016 PROCEEDING PDFDocument7 pagesSyafiqah ITC2016 PROCEEDING PDFFiqa SuccessNo ratings yet
- Environment AssignmentDocument2 pagesEnvironment AssignmentFiqa SuccessNo ratings yet
- Environment AssignmentDocument2 pagesEnvironment AssignmentFiqa SuccessNo ratings yet
- Acetobacter Xylinum Insertion With Inactivation of CelluloseDocument9 pagesAcetobacter Xylinum Insertion With Inactivation of CelluloseFiqa SuccessNo ratings yet
- PDF Edited RCEE 2016 IPVD PaperDocument6 pagesPDF Edited RCEE 2016 IPVD PaperFiqa SuccessNo ratings yet
- Mat101 w12 Hw6 SolutionsDocument8 pagesMat101 w12 Hw6 SolutionsKonark PatelNo ratings yet
- Reaffirmed 1998Document13 pagesReaffirmed 1998builconsNo ratings yet
- MICRF230Document20 pagesMICRF230Amador Garcia IIINo ratings yet
- The Hot Aishwarya Rai Wedding and Her Life.20130105.040216Document2 pagesThe Hot Aishwarya Rai Wedding and Her Life.20130105.040216anon_501746111100% (1)
- Hssive-Xi-Chem-4. Chemical Bonding and Molecular Structure Q & ADocument11 pagesHssive-Xi-Chem-4. Chemical Bonding and Molecular Structure Q & AArties MNo ratings yet
- Bethelhem Alemayehu LTE Data ServiceDocument104 pagesBethelhem Alemayehu LTE Data Servicemola argawNo ratings yet
- ELS 15 Maret 2022 REVDocument14 pagesELS 15 Maret 2022 REVhelto perdanaNo ratings yet
- ZEROPAY WhitepaperDocument15 pagesZEROPAY WhitepaperIlham NurrohimNo ratings yet
- Paper 11-ICOSubmittedDocument10 pagesPaper 11-ICOSubmittedNhat Tan MaiNo ratings yet
- E WiLES 2021 - BroucherDocument1 pageE WiLES 2021 - BroucherAshish HingnekarNo ratings yet
- Mod 2 MC - GSM, GPRSDocument61 pagesMod 2 MC - GSM, GPRSIrene JosephNo ratings yet
- Vermicomposting Learning ModulesDocument6 pagesVermicomposting Learning ModulesPamara Prema Khannae100% (1)
- Laminar Premixed Flames 6Document78 pagesLaminar Premixed Flames 6rcarpiooNo ratings yet
- Recent Advances in Dielectric-Resonator Antenna TechnologyDocument14 pagesRecent Advances in Dielectric-Resonator Antenna Technologymarceloassilva7992No ratings yet
- M 995Document43 pagesM 995Hossam AliNo ratings yet
- MODULE 8. Ceiling WorksDocument2 pagesMODULE 8. Ceiling WorksAj MacalinaoNo ratings yet
- First Semester-NOTESDocument182 pagesFirst Semester-NOTESkalpanaNo ratings yet
- ReflectionDocument3 pagesReflectionapi-174391216No ratings yet
- University Grading System - VTUDocument3 pagesUniversity Grading System - VTUmithilesh8144No ratings yet
- Th-Sunday Magazine 6 - 2Document8 pagesTh-Sunday Magazine 6 - 2NianotinoNo ratings yet
- Activity 4 - Energy Flow and Food WebDocument4 pagesActivity 4 - Energy Flow and Food WebMohamidin MamalapatNo ratings yet
- New Horizon Public School, Airoli: Grade X: English: Poem: The Ball Poem (FF)Document42 pagesNew Horizon Public School, Airoli: Grade X: English: Poem: The Ball Poem (FF)stan.isgod99No ratings yet